Installing R package: Fixing package ‘xxx’ is not available (for R version x.y.z)” warning?
The default version of `install.packages` somehow stopped working for me on latest version of R. In fact this is regardless of the version of R. I remember having same problem with the older versions of R as well.
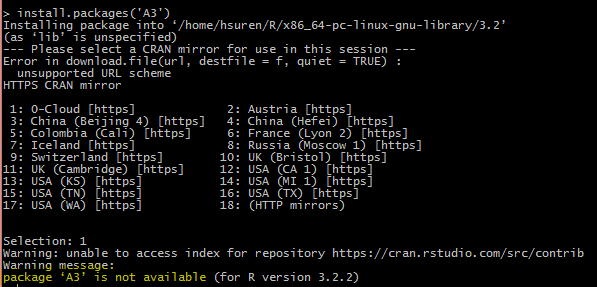
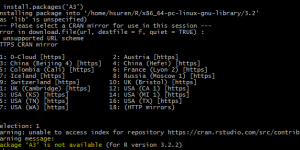
Here is the screen shot of the warning. if you are having similar problem, keep on reading.
Anyways, long story short, as everyone knows, if you want to install a package on R, you use:
install.packages('package-name')
And it asks you to pick a mirror repository to search the package in. If you are like me, picking the Cloud, you will soon realize, it will give you a misleading warning that the package is not available for your installed version of R. Well, this is not entirely true based on my experience.
I think not all the repositories include same packages (or versions) for some reason, that’s something I need to confirm it with R support team. If you select different mirror from previous step, and you are lucky enough, you might be able to install the package without any further problem/headache.
I usually use the following code to install package from R and it works 99% of the time.
install.packages('package-name',repos='http://cran.us.r-project.org')
There are several mirrors available you can try, simply change the repos field on the code above. Find the list of the mirrors here.
You still have problem installing the R package? See my other post here.
Feel free to share your experience, or the mirror works for you the best commenting below.







[…] of all, I highly recommend checking my other post if you are getting package ‘xxx’ is not available (for R version x.y.z)” […]
Thanks for the valuable suggestion. Your suggestion worked.
I am not able to install openNLP in R 3.6.2 .
ANY help would be appreciated 🙂
I am not being able to install plotly package,tried every method u given….what should i do to make it work.
Hi Leeza,
Can you share the R version and plotly version you are trying to install as well as the exact error message please? Thanks!
This was very helpful, thank you!
I tried install.packages(‘package-name’,repos=’http://cran.us.r-project.org’) but it didnt work, now I have a new warning Installing package into ‘/home/MYSELF/R/x86_64-pc-linux-gnu-library/3.2’
(as ‘lib’ is unspecified)
what does it mean the lib is unspecified
can you please copy the exact error in the console?
HI, i tried the link you gave but this is the respond i saw on the console
Warning message:
package ‘agricolae’ was built under R version 3.6.2
this was the command i ran
install.packages(‘agricolae’,repos=’http://cran.us.r-project.org’)
i am using r version 3.6.1
THanks a lot
I am also having problem in installing worldcloud and sentiment package in R. it gives an error.
Warning in install.packages :
package ‘worldcloud’ is not available (for R version 3.4.4)
Warning in install.packages :
package ‘sentiment’ is not available (for R version 3.4.4)
what can I do in this regard?
By trying this
install.packages(‘package-name’,repos=’http://cran.us.r-project.org’)
It gives me this error
Warning in install.packages :
unable to access index for repository http://cran.us.r-project.org/src/contrib:
cannot open URL ‘http://cran.us.r-project.org/src/contrib/PACKAGES’
Warning in install.packages :
package ‘worldcloud’ is not available (for R version 3.4.4)
Warning in install.packages :
unable to access index for repository http://cran.us.r-project.org/bin/windows/contrib/3.4:
cannot open URL ‘http://cran.us.r-project.org/bin/windows/contrib/3.4/PACKAGES’
Can you make sure the package name is correct. I think it should be “wordcloud” instead of “worldcloud”?
Hi,
I am having some kind of problem in R. I tried to install “varest” package however it looks unavaliable. Pls could you help me about this issue.
Here is the problem that I have.
> install.packages(‘varest’)
Installing package into ‘C:/Users/user/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘varest’ is not available (for R version 3.5.1)
Did you try?
install.packages(‘varest’,repos=’http://cran.us.r-project.org’)
Yes I tried also that code for different versions of R. Maybe I have to reinstall. I would miss something when I install program.
> install.packages(‘varest’,repos=’http://cran.us.r-project.org’)
Installing package into ‘C:/Users/user/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘varest’ is not available (for R version 3.5.1)
I’d try some more mirrors from here https://cran.r-project.org/mirrors.html.
Actually, varest is not the package name. It is a function in package called “sampling”
So please try this:
install.packages("sampling",repos="http://cran.us.r-project.org")Problem is solved. Thank you. I am really apricated.
package ‘deal’ is not available (for R version 3.5.1)
Hi, Suren!
I’m studying “Bayesian Network Credit Scoring in R”!
The code presented in related document uses libray “deal”.
When I installed package for deal, the following error occured!
Please help me!!!
=========================================
install.packages(‘deal’,repos=’http://cran.us.r-project.org’)
Installing package into ‘C:/Users/admin/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘deal’ is not available (for R version 3.5.1)
Seems like, `deal` package is removed from CRAN
You should try building it from source.
I tried to install the package lubridate. it show me below mentioned error
> install.packages(“lubridate”)
Warning in install.packages :
unable to access index for repository https://cran.rstudio.com/src/contrib:
cannot open URL ‘https://cran.rstudio.com/src/contrib/PACKAGES’
Installing package into ‘C:/Users/DELL/Documents/R/win-library/3.4’
(as ‘lib’ is unspecified)
Warning in install.packages :
unable to access index for repository https://cran.rstudio.com/src/contrib:
cannot open URL ‘https://cran.rstudio.com/src/contrib/PACKAGES’
Warning in install.packages :
package ‘lubridate’ is not available (for R version 3.4.3)
Warning in install.packages :
unable to access index for repository https://cran.rstudio.com/bin/windows/contrib/3.4:
cannot open URL ‘https://cran.rstudio.com/bin/windows/contrib/3.4/PACKAGES’
Did you try using a mirror like “http://cran.us.r-project.org”?
Hi Team,
I am getting the error while installing forecast package as mention below.
> install.packages(‘forecast’,repos=’http://cran.us.r-project.org’)
Warning: unable to access index for repository http://cran.us.r-project.org/src/contrib:
cannot open URL ‘http://cran.us.r-project.org/src/contrib/PACKAGES’
Warning: unable to access index for repository http://cran.us.r-project.org/bin/windows/contrib/3.4:
cannot open URL ‘http://cran.us.r-project.org/bin/windows/contrib/3.4/PACKAGES’
Warning message:
package ‘forecast’ is not available (for R version 3.4.1) .
Did you try other mirrors? https://cran.r-project.org/mirrors.html. Try the one closest to your location.
No, Now Getting error like install.packages(‘forecast’,repos=’https://cran.r-project.org/mirrors.html’)
Error: unexpected input in “install.packages(‘”
Hi Suren,
I am having some kind of problems in R.
I tried to install “GeneR” package however it looks unavaliable.Could you help me about this issue? I also try the above methods,but it can’t solve my problems.
Here is the problem that I have.
> install.packages(“GeneR”,repos=”http://cran.us.r-project.org”)
Installing package into ‘/home/xinw/R/x86_64-pc-linux-gnu-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘GeneR’ is not available (for R version 3.5.1)
I have taken one day to solve this problem,so I look forward to you reply.
Thank you very much!
It is because GeneR is on Bioconductor.
Try this:
source(“http://bioconductor.org/biocLite.R”)
biocLite(“GeneR”)
#and after succesfull install
library(GeneR)
It has some other problems.
> source(‘http://bioconductor.org/biocLite.R’)
Bioconductor version 3.7 (BiocInstaller 1.30.0), ?biocLite for help
> biocLite(‘GeneR’)
BioC_mirror: https://bioconductor.org
Using Bioconductor 3.7 (BiocInstaller 1.30.0), R 3.5.1 (2018-07-02).
Installing package(s) ‘GeneR’
installation path not writeable, unable to update packages: MASS
Warning message:
package ‘GeneR’ is not available (for R version 3.5.1)
Make sure you have write access to `.libPaths()` for the user you are using. From the error message, it seems like you do not have write access to that folder.
I change the access to ‘.libPaths()’:
# chmod a+w /usr/local/lib/R/site-library
# chmod a+w /usr/lib/R/site-library
# chmod a+w /usr/lib/R/library
> biocLite(‘GeneR’)
BioC_mirror: https://bioconductor.org
Using Bioconductor 3.7 (BiocInstaller 1.30.0), R 3.5.1 (2018-07-02).
Installing package(s) ‘GeneR’
Old packages: ‘MASS’
Update all/some/none? [a/s/n]:
a
trying URL ‘https://cloud.r-project.org/src/contrib/MASS_7.3-51.tar.gz’
Content type ‘application/x-gzip’ length 487219 bytes (475 KB)
==================================================
downloaded 475 KB
* installing *source* package ‘MASS’ …
** package ‘MASS’ successfully unpacked and MD5 sums checked
mv: cannot move ‘/usr/lib/R/library/MASS’ to ‘/usr/lib/R/library/00LOCK-MASS/MASS’: Permission denied
Warning in file.copy(f, instdir, TRUE) :
problem copying ./NAMESPACE to /usr/lib/R/library/MASS/NAMESPACE: Permission denied
Warning in file(file, if (append) “a” else “w”) :
cannot open file ‘/usr/lib/R/library/MASS/DESCRIPTION’: Permission denied
Error in file(file, if (append) “a” else “w”) :
cannot open the connection
ERROR: installing package DESCRIPTION failed for package ‘MASS’
* removing ‘/usr/lib/R/library/MASS’
The downloaded source packages are in
‘/tmp/RtmpL6NtAU/downloaded_packages’
Updating HTML index of packages in ‘.Library’
Warning messages:
1: package ‘GeneR’ is not available (for R version 3.5.1)
2: In install.packages(update[instlib == l, “Package”], l, contriburl = contriburl, :
installation of package ‘MASS’ had non-zero exit status
3: In file.create(f.tg) :
cannot create file ‘/usr/share/R/doc/html/packages.html’, reason ‘Permission denied’
4: In make.packages.html(.Library) : cannot update HTML package index
There are more problems,I am a R beginner.
I don‘t konw how to do that.I look forward to you reply.
Thank you very much.
Seems like it is still user write problem for the given directory.
If you are using R in command line, can you try running it with sudo?
sudo R –vanilla
and then install the packages…
Let me know how it goes.
> biocLite(“GeneR”)
BioC_mirror: https://bioconductor.org
Using Bioconductor 3.7 (BiocInstaller 1.30.0), R 3.5.1 (2018-07-02).
Installing package(s) ‘GeneR’
Old packages: ‘MASS’
Update all/some/none? [a/s/n]:
n
Warning message:
package ‘GeneR’ is not available (for R version 3.5.1)
i’d try downloading the source and install it manually http://www.bioconductor.org/packages/2.3/bioc/html/GeneR.html
I’d try running R with sudo.
~$ sudo R -vanilla
> source(“http://bioconductor.org/biocLite.R”)
Bioconductor version 3.7 (BiocInstaller 1.30.0), ?biocLite for help
> biocLite(“GeneR”)
BioC_mirror: https://bioconductor.org
Using Bioconductor 3.7 (BiocInstaller 1.30.0), R 3.5.1 (2018-07-02).
Installing package(s) ‘GeneR’
Old packages: ‘MASS’
Update all/some/none? [a/s/n]: a
trying URL ‘https://cloud.r-project.org/src/contrib/MASS_7.3-51.tar.gz’
Content type ‘application/x-gzip’ length 487219 bytes (475 KB)
==================================================
downloaded 475 KB
* installing *source* package ‘MASS’ …
** package ‘MASS’ successfully unpacked and MD5 sums checked
** libs
gcc -std=gnu99 -I”/usr/share/R/include” -DNDEBUG -fpic -g -O2 -fdebug-prefix-map=/build/r-base-oNcpyf/r-base-3.5.1=. -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -g -c MASS.c -o MASS.o
gcc -std=gnu99 -I”/usr/share/R/include” -DNDEBUG -fpic -g -O2 -fdebug-prefix-map=/build/r-base-oNcpyf/r-base-3.5.1=. -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -g -c lqs.c -o lqs.o
gcc -std=gnu99 -shared -L/usr/lib/R/lib -Wl,-Bsymbolic-functions -Wl,-z,relro -o MASS.so MASS.o lqs.o -L/usr/lib/R/lib -lR
installing to /usr/lib/R/library/MASS/libs
** R
** data
*** moving datasets to lazyload DB
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** testing if installed package can be loaded
* DONE (MASS)
The downloaded source packages are in
‘/tmp/RtmpIdmoWh/downloaded_packages’
Updating HTML index of packages in ‘.Library’
Making ‘packages.html’ … done
Warning message:
package ‘GeneR’ is not available (for R version 3.5.1)
But there is the same problem.How could I do?
Look forward your reply.Thank you very much.
did you try installing the package from source?
Go here and download tar.gz file https://bioc.ism.ac.jp/packages/2.8/bioc/html/GeneR.html
And then in the terminal run
`R CMD INSTALL GeneR_2.22.0.tar.gz`
it should work
Thanks for your reply.
I install the package from source and run the command in the terminal.
But I think my OS is Ubuntu,it can’t work.
:~/R/x86_64-pc-linux-gnu-library/3.5$ R cmd install GeneR_2.22.0.tar.gz
ARGUMENT ‘cmd’ __ignored__
ARGUMENT ‘install’ __ignored__
ARGUMENT ‘GeneR_2.22.0.tar.gz’ __ignored__
And the information of R is:
R version 3.5.1 (2018-07-02) — “Feather Spray”
Copyright (C) 2018 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit).
And the IDE of R is R-studio.
So is it the problem of my OS?
Look forward to your reply,thanks a lot!
Please copy from my code, the command should be all upper case.
:~/R/x86_64-pc-linux-gnu-library/3.5$ R CMD INSTALL GeneR_2.22.0.tar.gz
* installing to library ‘/home/R/x86_64-pc-linux-gnu-library/3.5’
* installing *source* package ‘GeneR’ …
** libs
g++ -I”/usr/share/R/include” -DNDEBUG -fpic -g -O2 -fdebug-prefix-map=/build/r-base-oNcpyf/r-base-3.5.1=. -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -g -c GeneR_glob.cc -o GeneR_glob.o
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h: In member function ‘void GeneR_seq::setAccN(char*)’:
GeneR_seq.h:156:15: error: no match for ‘operator<<’ (operand types are ‘std::basic_ostream’ and ‘std::ostream {aka std::basic_ostream}’)
std::cout << "GeneR::GeneR_seq::setAccN Ouch name "<<_id<<
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
"too long" << std::cout;
~~~~~~~~~~~^~~~~~
GeneR_seq.h:156:15: note: candidate: operator<<(int, int)
GeneR_seq.h:156:15: note: no known conversion for argument 2 from ‘std::ostream {aka std::basic_ostream}’ to ‘int’
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:108:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(std::basic_ostream::__ostream_type& (*)(std::basic_ostream::__ostream_type&)) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(__ostream_type& (*__pf)(__ostream_type&))
^~~~~~~~
/usr/include/c++/7/ostream:108:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘std::basic_ostream::__ostream_type& (*)(std::basic_ostream::__ostream_type&) {aka std::basic_ostream& (*)(std::basic_ostream&)}’
/usr/include/c++/7/ostream:117:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(std::basic_ostream::__ios_type& (*)(std::basic_ostream::__ios_type&)) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream; std::basic_ostream::__ios_type = std::basic_ios]
operator<<(__ios_type& (*__pf)(__ios_type&))
^~~~~~~~
/usr/include/c++/7/ostream:117:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘std::basic_ostream::__ios_type& (*)(std::basic_ostream::__ios_type&) {aka std::basic_ios& (*)(std::basic_ios&)}’
/usr/include/c++/7/ostream:127:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(std::ios_base& (*)(std::ios_base&)) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(ios_base& (*__pf) (ios_base&))
^~~~~~~~
/usr/include/c++/7/ostream:127:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘std::ios_base& (*)(std::ios_base&)’
/usr/include/c++/7/ostream:166:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(long int) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(long __n)
^~~~~~~~
/usr/include/c++/7/ostream:166:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘long int’
/usr/include/c++/7/ostream:170:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(long unsigned int) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(unsigned long __n)
^~~~~~~~
/usr/include/c++/7/ostream:170:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘long unsigned int’
/usr/include/c++/7/ostream:174:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(bool) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(bool __n)
^~~~~~~~
/usr/include/c++/7/ostream:174:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘bool’
In file included from /usr/include/c++/7/ostream:693:0,
from /usr/include/c++/7/iostream:39,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/bits/ostream.tcc:91:5: note: candidate: std::basic_ostream& std::basic_ostream::operator<<(short int) [with _CharT = char; _Traits = std::char_traits]
basic_ostream::
^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
/usr/include/c++/7/bits/ostream.tcc:91:5: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘short int’
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:181:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(short unsigned int) [with_CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(unsigned short __n)
^~~~~~~~
/usr/include/c++/7/ostream:181:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘short unsigned int’
In file included from /usr/include/c++/7/ostream:693:0,
from /usr/include/c++/7/iostream:39,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/bits/ostream.tcc:105:5: note: candidate: std::basic_ostream& std::basic_ostream::operator<<(int) [with _CharT = char; _Traits = std::char_traits]
basic_ostream::
^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
/usr/include/c++/7/bits/ostream.tcc:105:5: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘int’
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:192:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(unsigned int) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(unsigned int __n)
^~~~~~~~
/usr/include/c++/7/ostream:192:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘unsigned int’
/usr/include/c++/7/ostream:201:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(long long int) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(long long __n)
^~~~~~~~
/usr/include/c++/7/ostream:201:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘long long int’
/usr/include/c++/7/ostream:205:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(long long unsigned int) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(unsigned long long __n)
^~~~~~~~
/usr/include/c++/7/ostream:205:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘long long unsigned int’
/usr/include/c++/7/ostream:220:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(double) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(double __f)
^~~~~~~~
/usr/include/c++/7/ostream:220:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘double’
/usr/include/c++/7/ostream:224:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(float) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(float __f)
^~~~~~~~
/usr/include/c++/7/ostream:224:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘float’
/usr/include/c++/7/ostream:232:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(long double) [with _CharT= char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(long double __f)
^~~~~~~~
/usr/include/c++/7/ostream:232:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘long double’
/usr/include/c++/7/ostream:245:7: note: candidate: std::basic_ostream::__ostream_type& std::basic_ostream::operator<<(const void*) [with _CharT= char; _Traits = std::char_traits; std::basic_ostream::__ostream_type = std::basic_ostream]
operator<<(const void* __p)
^~~~~~~~
/usr/include/c++/7/ostream:245:7: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘const void*’
In file included from /usr/include/c++/7/ostream:693:0,
from /usr/include/c++/7/iostream:39,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/bits/ostream.tcc:119:5: note: candidate: std::basic_ostream& std::basic_ostream::operator<<(std::basic_ostream::__streambuf_type*) [with _CharT = char; _Traits = std::char_traits; std::basic_ostream::__streambuf_type = std::basic_streambuf]
basic_ostream::
^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
/usr/include/c++/7/bits/ostream.tcc:119:5: note: no known conversion for argument 1 from ‘std::ostream {aka std::basic_ostream}’ to ‘std::basic_ostream::__streambuf_type* {aka std::basic_streambuf*}’
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:682:5: note: candidate: template typename std::enable_if<std::__and_<std::__not_<std::is_lvalue_reference >, std::__is_convertible_to_basic_ostream, std::__is_insertable<typename std::__is_convertible_to_basic_ostream::__ostream_type, const _Tp&, void> >::value, typename std::__is_convertible_to_basic_ostream::__ostream_type>::type std::operator<<(_Ostream&&, const _Tp&)
operator<<(_Ostream&& __os, const _Tp& __x)
^~~~~~~~
/usr/include/c++/7/ostream:682:5: note: template argument deduction/substitution failed:
/usr/include/c++/7/ostream: In substitution of ‘template typename std::enable_if<std::__and_<std::__not_<std::is_lvalue_reference >, std::__is_convertible_to_basic_ostream, std::__is_insertable<typename std::__is_convertible_to_basic_ostream::__ostream_type, const _Tp&, void> >::value, typename std::__is_convertible_to_basic_ostream::__ostream_type>::type std::operator<<(_Ostream&&, const _Tp&) [with _Ostream = std::basic_ostream&; _Tp = std::basic_ostream]’:
GeneR_seq.h:156:23: required from here
/usr/include/c++/7/ostream:682:5: error: no type named ‘type’ in ‘struct std::enable_if<false, std::basic_ostream&>’
/usr/include/c++/7/ostream:574:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const unsigned char*)
operator<<(basic_ostream& __out, const unsigned char* __s)
^~~~~~~~
/usr/include/c++/7/ostream:574:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘const unsigned char*’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:569:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const signed char*)
operator<<(basic_ostream& __out, const signed char* __s)
^~~~~~~~
/usr/include/c++/7/ostream:569:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘const signed char*’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:556:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const char*)
operator<<(basic_ostream& __out, const char* __s)
^~~~~~~~
/usr/include/c++/7/ostream:556:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘const char*’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/ostream:693:0,
from /usr/include/c++/7/iostream:39,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/bits/ostream.tcc:321:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const char*)
operator<<(basic_ostream& __out, const char* __s)
^~~~~~~~
/usr/include/c++/7/bits/ostream.tcc:321:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘const char*’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:539:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const _CharT*)
operator<<(basic_ostream& __out, const _CharT* __s)
^~~~~~~~
/usr/include/c++/7/ostream:539:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: mismatched types ‘const _CharT*’ and ‘std::basic_ostream’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:519:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, unsigned char)
operator<<(basic_ostream& __out, unsigned char __c)
^~~~~~~~
/usr/include/c++/7/ostream:519:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘unsigned char’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:514:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, signed char)
operator<<(basic_ostream& __out, signed char __c)
^~~~~~~~
/usr/include/c++/7/ostream:514:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘signed char’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:508:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, char)
operator<<(basic_ostream& __out, char __c)
^~~~~~~~
/usr/include/c++/7/ostream:508:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘char’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:502:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, char)
operator<<(basic_ostream& __out, char __c)
^~~~~~~~
/usr/include/c++/7/ostream:502:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘char’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/iostream:39:0,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/ostream:497:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, _CharT)
operator<<(basic_ostream& __out, _CharT __c)
^~~~~~~~
/usr/include/c++/7/ostream:497:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: deduced conflicting types for parameter ‘_CharT’ (‘char’ and ‘std::basic_ostream’)
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/bits/ios_base.h:46:0,
from /usr/include/c++/7/ios:42,
from /usr/include/c++/7/ostream:38,
from /usr/include/c++/7/iostream:39,
from GeneR_seq.h:18,
from GeneR_glob.cc:13:
/usr/include/c++/7/system_error:217:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const std::error_code&)
operator<<(basic_ostream& __os, const error_code& __e)
^~~~~~~~
/usr/include/c++/7/system_error:217:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: cannot convert ‘std::cout’ (type ‘std::ostream {aka std::basic_ostream}’) to type ‘const std::error_code&’
“too long” << std::cout;
^~~~
In file included from /usr/include/c++/7/string:52:0,
from GeneR_seq.h:15,
from GeneR_glob.cc:13:
/usr/include/c++/7/bits/basic_string.h:6275:5: note: candidate: template std::basic_ostream& std::operator<<(std::basic_ostream&, const std::__cxx11::basic_string&)
operator<<(basic_ostream& __os,
^~~~~~~~
/usr/include/c++/7/bits/basic_string.h:6275:5: note: template argument deduction/substitution failed:
In file included from GeneR_glob.cc:13:0:
GeneR_seq.h:156:23: note: ‘std::ostream {aka std::basic_ostream}’ is not derived from ‘const std::__cxx11::basic_string’
“too long” << std::cout;
^~~~
/usr/lib/R/etc/Makeconf:166: recipe for target 'GeneR_glob.o' failed
make: *** [GeneR_glob.o] Error 1
ERROR: compilation failed for package ‘GeneR’
* removing ‘/home/R/x86_64-pc-linux-gnu-library/3.5/GeneR’
Mr Haktan Suren,there is the new problem.
Should i try to reinstall R ?
Because it still have the problem,I don't know how to do,i lost my confidence.
Thank you very much for your patience.
Look forward to your reply again!
No I do not think R reinstall would fix this. You are getting a compilation error. Btw, May I ask why you want to use GeneR package? Maybe you can find some other alternative?
Thanks for your reply.
I am a beginner of the R language.
Currently I want to process a .fa file and need to use the readFasta() function.
I mainly use R for the analysis of biological information data.
I consulted R’s book and said that there are two ways to install R. An installation release, a compilation and installation from scratch. And it is recommended to use the release version for installation. I am thinking about whether I am compiling and installing from scratch, so it will be more troublesome in the subsequent work.
I install other packages also have some problems.And it wasted me a lot time.
So I am thinking about reinstalling R.
Then go ahead and reinstall the R. And try to install any package.
Thank you for your reply.
I install the package of ‘GeneR’ successfully.
I download the ‘GeneR’ package from ‘https://www.bioconductor.org/help/search/index.html?q=GeneR/’
Then I install from Package Archive File(.tar.gz):
~$ tar -xzf /R/x86_64-pc-linux-gnu-library/3.5/GeneR_1.4.2.tar.gz
~$ cd /home/xinw/R/x86_64-pc-linux-gnu-library/3.5/GeneR
~/R/x86_64-pc-linux-gnu-library/3.5/GeneR$ echo ‘exportPattern( “.” )’ > NAMESPACE
~/R/x86_64-pc-linux-gnu-library/3.5/GeneR$ cd ..
~/R/x86_64-pc-linux-gnu-library/3.5$ tar -zcf GeneR_1.4.2.tar.gz GeneR
~/R/x86_64-pc-linux-gnu-library/3.5$ R CMD INSTALL GeneR_1.4.2.tar.gz
……
* DONE (GeneR)
> library(“GeneR”, lib.loc=”~/R/x86_64-pc-linux-gnu-library/3.5″)
> readFasta(“chrY.fa”)
Error in .C(“sys_free”, as.integer(seqno), PACKAGE = “GeneR”) :
“sys_free” not available for .C() for package “GeneR”
But there is the new problem.
Hi,
I tried to install ggrepe1 using – install.packages(‘ggrepe1′,repos=’http://cran.us.r-project.org’)
it’s showing me this error – package ‘ggrepe1’ is not available (for R version 3.5.1)
I also tried to update my R, but it says that i’m using the latest version of R. How can i install the package now ?
Thanks in Advance.
Maybe a typo in the package name? try install.packages(‘ggrepel′,repos=’http://cran.us.r-project.org’)
Hi Suren,
I have a problems in R.
I tried to install “gplots” package but it’s seems i can’t access to a cran miror .Could you help me about this issue? I also try the above methods,but it can’t solve my problem.
i also try other packages, but i have the same problem
i have R VERSION 3.5.2 i switch to an other version (2.10.1) ,with this version i can instal packages but i can’t find the package gplots
so, please what can i do to solve this problem?
Hi Suren,
I have a problem in R.
I tried to install “gplots” package but without succes .Could you help me about this issue? I also tried the above methods,but it can’t solve my problem.
i aloso tried other packages , i have the same problem
i use R version 3.5.2 and when i try other version like version 2.10.1 i haven’t find the “gplots” package so i tried other packages like geneR and this time it’s work but i don’t find gplots package in the version 2.10.1
so ,please, what i have to do solve this problem
Please try one of the mirrors from https://cran.r-project.org/mirrors.html close to your location. Let us know if it works
Hi!
I am also trying to create “Beautiful” result tables for my panel data models using the sjPlot package and its sjt.lm function as mentioned in the following link:
https://www.r-bloggers.com/beautiful-tables-for-linear-model-summaries-rstats/
but none of them works. I get the following error messages:
> model1 install.packages(“sjt.lm”)
Installing package into ‘C:/Users/Sinem/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘sjt.lm’ is not available (for R version 3.5.2)
> install.packages(‘sjt.lm’,repos=’http://cran.us.r-project.org’)
Installing package into ‘C:/Users/Sinem/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘sjt.lm’ is not available (for R version 3.5.2)
How can I solve the problem? Or could you please suggest me any alternatives (as simple as possible since I am a super beginner only, like I have no clue what “mirroring” is) so that I can create result tables with coefficients for my models that I can present? I don’t mind which package / function as long as it works.
Thank you so much in advance!
Hi,
The package name is `sjPlot` so
try running
install.packages("sjPlot"). It should work!
Hi
I am trying to install the ‘demography’ package in Mac OS Mojave 10.14 (18A391) and I get the following error Warning in install.packages:
package ‘demography’ is not available (for R version 3.3.2) my version is RStudio
Version 1.1.463 – © 2009-2018 RStudio, Inc
Yes it is a correct warning, package requires at least 3.4 version of R. Please upgrade the R and try to upload the package.
Hi
I am trying to install the ‘rgp’ package and I get the following error Warning in install.packages:
package ‘rgp’ is not available (for R version 3.5.1)
Thank you in advance
Probably due the package is removed from CRAN https://cran.r-project.org/web/packages/rgp/index.html. You have to install it manually.
Hi
I am trying to install the ‘xgboost’ package and I get the following error Warning in install.packages:
package ‘xgboost’ is not available (for R version 3.2.2)
I tried:
1. install.packages(“drat”, repos=”https://cran.rstudio.com”)
drat:::addRepo(“dmlc”)
install.packages(“xgboost”, repos=”http://dmlc.ml/drat/”, type = “source”)
2. install.packages(“xgboost”)
3. install.packages(‘xgboost’,repos=’http://cran.us.r-project.org’)
Unfortunately, I did not make progress.
Thank you in advance
Hi Luciana,
I think it is correct error. xgboost depends R (>= 3.3.0). You need to upgrade your R version to be able to install the package.
How to install ggradar
seems like it is part of `ggiraphExtra`. Go ahead and try install.packages(“ggiraphExtra”)
> install.packages(“pandas”,repos=”http://cran.us.r-project.org”)
package ‘pandas’ is not available (for R version 3.4.3)
are you sure there is a package named “pandas” i could not find it in Google.
https://pandas.pydata.org
well, it is not an R package. It’s a python package. That’s why you can’t install it.
Hi I cannot install IpSolve. This is the message I get:
install.packages(“IpSolve”, repos=”https://cran.ma.imperial.ac.uk/”)
Installing package into ‘/Users/maria/Library/R/3.5/library’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘IpSolve’ is not available (for R version 3.5.2)
I tried also IpSolveAPI, and all the links from my country in the link that you posted!
Any ideas?
thanks in advance!
hi, i’ve solved this thanks!
Hi Maria:
I’ve been using Windows and my R version is 3.4.2 I am having same problem as you wrote first. But my package is different. However, what the technique did you follow?
When I try to install package ‘openNLPmodels.en’ in RStudio I get the following error messages: I run the latest versions of RStudio an R. openNLP and openNLPdata are correctly installed. please help me install it.
install.packages(“openNLPmodels.en”, dependencies=TRUE, repos =
“http://datacube.wu.ac.at/”, type = “source”, INSTALL_opts = c(‘–no-lock’))
Warning in install.packages :
cannot open URL ‘http://datacube.wu.ac.at/src/contrib/PACKAGES.rds’: HTTP status was ‘404 Not Found’
trying URL ‘http://datacube.wu.ac.at/src/contrib/openNLPmodels.en_1.5-1.tar.gz’
Content type ‘application/x-gzip’ length 77794529 bytes (74.2 MB)
downloaded 74.2 MB
‘\\WWG00M.ROOTDOM.NET\DFS\HOME\wt2rrib’
CMD.EXE was started with the above path as the current directory.
UNC paths are not supported. Defaulting to Windows directory.
ERROR: unable to create ‘\\Wwg00m.rootdom.net/dfs/HOME/WT2RRIB/R/Sentimental Analysis/openNLPmodels.en’
In R CMD INSTALL
Warning in install.packages :
installation of package ‘openNLPmodels.en’ had non-zero exit status
From the error message, it seems like you do not have write permission for the directory. You may want to try running R as an administrator and then try to install the package. Let us know if it works.
Thanks for replying.
I am already using R and downloaded many packages already but facing problem in downloading this particular package only
i have a problem to install package PoEdata
install.packages(“devtools”) # if not already installed
library(devtools)
install_git(“https://github.com/ccolonescu/PoEdata”)
have you tried these from https://github.com/ccolonescu/PoEdata
package ‘devtools’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\hp\AppData\Local\Temp\RtmpuQCbR3\downloaded_packages
> library(“devtools”)
Error: package or namespace load failed for ‘devtools’ in loadNamespace(j install_git(“https://github.com/ccolonescu/PoEdata”)
Error: unexpected input in “install_git(“”
> install_git(“https://github.com/ccolonescu/PoEdata”)
Error in install_git(“https://github.com/ccolonescu/PoEdata”) :
could not find function “install_git”
Can you try restarting R and loading the devtools library again?
Hi!Suren.
there is my problem:
> install.packages(“yitian”)
Installing package into ‘C:/Users/win/Documents/R/win-library/3.5’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘yitian’ is not available (for R version 3.5.1)
please help me? thank you very much!
Are you sure about the package name? Where did you find it?
Hi
Any help for installing “rscproxy” to R version 3.5.1.
Thank you!
I think you should install manually since it says it is archived https://cran.r-project.org/web/packages/rscproxy/index.html.
Hello,
Could you advise me on what to install to fix
package ‘FactMineR’ is not available (for R version 3.4.4)
My best, Keke
Maybe you mean “FactoMineR”? https://cran.r-project.org/web/packages/FactoMineR/index.html
Dear Haktan Suren,
Could you please help my case as i tried to installed fARMA but it failed even i tried to download manually but it showed the status:
fArma_3010.79.tar.gz’ had non-zero exit status
thank you so much in advance for you help.
seems like is removed from R repository https://cran.r-project.org/web/packages/fArma/index.html
You need to install it from source
how can I install it from source?
you can find the info here https://support.rstudio.com/hc/en-us/articles/218004217-Building-R-from-source
hello sir Haktan Suren,
when i try to install ggplot2 : install.packages (‘ggplot2’)
i heve this error:
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
— SVP sélectionner un miroir CRAN pour cette session —
Avis : impossible d’accéder à l’index de l’entrepôt https://cloud.r-project.org/src/contrib
Message d’avis :
package ‘ggplot2’ is not available (for R version 3.0.2)
help please
you should upgrade your R version. ggplot2 has R (≥ 3.1) dependency.
Thank you sir, after that and when try to execute my code i have this error:
Warning message:
fermeture de la connexion inutilisée 3 (results/figure-5-retx-count/car-relay-tx.txt.bz2)
Warning message:
fermeture de la connexion inutilisée 3 (results/figure-5-retx-count/car-relay-in-cache.txt.bz2)
Warning messages:
1: `geom_bar()` no longer has a `binwidth` parameter. Please use `geom_histogram()` instead.
2: `axis.ticks.margin` is deprecated. Please set `margin` property of `axis.text` instead
3: `legend.margin` must be specified using `margin()`. For the old behavior use legend.spacing
Erreur : stat_bin() must not be used with a y aesthetic.
Exécution arrêtée
sir, i am not being able to install readr package. it keeps on showing unable to open url, package is not available for r version 3.5.2.
Did you try the other mirrors from here https://cran.r-project.org/mirrors.html?
Hi. I read through the above questions and attempted to your suggestions. However, no luck; I am probably missing something. I am trying to use the package car though it will not install due version incompatibility. The error message is pasted below. Also Rtools package received the same message–not available for 3.5. Do you have any suggestions? Best. Frances
> install.packages(“car”)
WARNING: Rtools is required to build R packages but is not currently installed. Please download and install the appropriate version of Rtools before proceeding:
https://cran.rstudio.com/bin/windows/Rtools/
Installing package into ‘C:/Users/Frances/Documents/R/win-library/3.4’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘car’ is not available (for R version 3.4.1)
Do you have the Rtools installed from here https://cran.r-project.org/bin/windows/Rtools/
I appreciate your reply. I installed Rtools as you recommended. However, I receive the same error as last time (see below). I would like to use ‘car’ as it was recommended that this package works well with unbalanced designs. Do you have any suggestions on how to install ‘car’ or an alternative to ‘car’? Thanks for the help. I will keep investigating this error also. Best. Frances
install.packages(“car”)
Installing package into ‘C:/Users/Frances/Documents/R/win-library/3.4’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘car’ is not available (for R version 3.4.1)
well it seems like you need to upgrade R version to 3.5.0 to use car R (≥ 3.5.0)
Hi There. Thanks for the post and all information. I still have problems to install pdftools on my R (v3.2.2) Services for SQLServer, because can’t find the library. I tested a lot of repositories without success.
Now i tryed installer manually using a local file (not repository) and have no problems for install, but now appears a error message every time i tryed to use the library with then simple command: “library(pdftools)”
The error is:
Error in readRDS(pfile) :
cannot read workspace version 3 written by R 3.6.0; need R 3.5.0 or newer
Calls: source … library -> find.package -> lapply -> FUN -> readRDS
what is your R version? ever tried upgrading it?
Dear all…
please help me i am much frustrated now because of i am trying to install package INLA, but every time i receiving following error/warning messages,i am using R version 3.6.2:
Error in read.dcf(file.path(pkgname, “DESCRIPTION”), c(“Package”, “Type”)) :
cannot open the connection
In addition: Warning messages:
1: In download.file(url, destfile, method, mode = “wb”, …) :
downloaded length 210960384 != reported length 335976807
2: In unzip(zipname, exdir = dest) : error 1 in extracting from zip file
3: In read.dcf(file.path(pkgname, “DESCRIPTION”), c(“Package”, “Type”)) :
cannot open compressed file ‘INLA/DESCRIPTION’, probable reason ‘No such file or directory’
Inaddition when i install package “DESCRIPTION”, i received error:
Installing package into ‘C:/Users/Dell/Documents/R/win-library/3.6’
(as ‘lib’ is unspecified)
Warning message:
package ‘DESCRIPTION’ is not available (for R version 3.6.2)
Error says it does not find the file name “DESCRIPTION” in the directory. This is not a package issue.
i have tried following codes, but unable to install and above error message generated,
install.packages(“INLA”, repos=c(getOption(“repos”), INLA=”https://inla.r-inla-download.org/R/stable”), dep=TRUE)
install.packages(“INLA”, repos=c(getOption(“repos”), INLA=”https://inla.r-inla-download.org/R/testing”), dep=TRUE)
Hi,
is there a way to find the install packages installed on a specific day and then delete them in one go?
kindly help
Hi!
I have problems installing the ‘dslabs’ package from CRAM.
The error message is the following one:
> install.packages(‘dslabs’)
Installing package into ‘/Users/Clara/Library/R/3.3/library’
(as ‘lib’ is unspecified)
also installing the dependency ‘ggplot2’
There are binary versions available but the source versions are later:
binary source needs_compilation
ggplot2 2.2.1 3.3.0 FALSE
dslabs 0.3.3 0.7.3 FALSE
installing the source packages ‘ggplot2’, ‘dslabs’
probando la URL ‘http://cran.rstudio.com/src/contrib/ggplot2_3.3.0.tar.gz’
Content type ‘unknown’ length 3031461 bytes (2.9 MB)
==================================================
downloaded 2.9 MB
probando la URL ‘http://cran.rstudio.com/src/contrib/dslabs_0.7.3.tar.gz’
Content type ‘unknown’ length 3388394 bytes (3.2 MB)
==================================================
downloaded 3.2 MB
* installing *source* package ‘ggplot2’ …
** package ‘ggplot2’ successfully unpacked and MD5 sums checked
** R
** data
*** moving datasets to lazyload DB
** inst
** preparing package for lazy loading
Error : object ‘glue_collapse’ is not exported by ‘namespace:glue’
ERROR: lazy loading failed for package ‘ggplot2’
* removing ‘/Users/Clara/Library/R/3.3/library/ggplot2’
Warning in install.packages :
installation of package ‘ggplot2’ had non-zero exit status
ERROR: dependency ‘ggplot2’ is not available for package ‘dslabs’
* removing ‘/Users/Clara/Library/R/3.3/library/dslabs’
Warning in install.packages :
installation of package ‘dslabs’ had non-zero exit status
The downloaded source packages are in
‘/private/var/folders/55/s42x1y8s5yngpvr00v_zpvw00000gn/T/RtmpkfZcBO/downloaded_packages’
could you help me please?
many thanks in advance!
Try updating the version of glue package. `install.packages(“glue”)`
Hi,
Thanks for sharing your tips.
Unfortunately I didn’t manage to resolve my ‘package ‘rpart.plot’ is not available (for R version 3.3.3)’ issue using various mirrors. I need to use the prp() function and can’t without the rpart.plot package…
I am using a Mac OS 10.10.5, and R version 3.3.3, R Studio 1.1.463. Could you please help me?
Thanks a million.
Alex
Resolved. I downloaded an older version of rpart, specifying it in the command
> install_version(‘rpart.plot’, version = ‘3.0.0’, repos = “http://cran.us.r-project.org”). Cheers, Alex
Thanks a lot.
Hi,
I have been trying to install shiny package on R 3.2.2(2015) on Linux server, so that i can run the shiny app R Code.
1_ it keeps giving error for packages not found “httpuv”,”promises”,”later”
2) Then i tried to install “later”,
3)I am able to install install.packages(“lattice”,”/home/agarwalm/R/”,repos=”http://cran.r-project.org”)
Hi,
I have been trying to install shiny package on R 3.2.2(2015) on Linux server, so that i can run the shiny app R Code.
1_ it keeps giving error for packages not found “httpuv”,”promises”,”later”
2) Then i tried to install “later”,
3)I am able to install install.packages(“lattice”,”/home/agarwalm/R/”,repos=”http://cran.r-project.org”)
4) getting this error when trying to install “lattice”
on “linux” Warning: unable to access index for repository http://cran.case.edu/src/contrib
Try different mirror sites from here https://cran.r-project.org/mirrors.html
hey there.i tried both the way yours and the traditional one but it still not installing..help!
follwing is copied message that i get. plss see into this ;
> install.packages(‘Agreement’,repos=’http://cran.us.r-project.org’)
Installing package into ‘C:/Users/ANJALI/Documents/R/win-library/3.6’
(as ‘lib’ is unspecified)
Warning message:
package ‘Agreement’ is not available (for R version 3.6.3)
>
I guess you have to install that package manually. It is removed from the CRAN repository. See the link https://cran.r-project.org/web/packages/Agreement/index.html
Hi, thanks for your sharing. However, I tried to use your method to install the package ‘ROCR’, but it did not work. The warning message is:
Warning in install.packages :
package ‘ROCR’ is not available (for R version 3.5.1)
May I know how to solve this? Thank you.
Hi, thanks for the sharing. However, I tried to use your method to install the package ‘ROCR’, but it did not work. The result is:
Warning in install.packages :
package ‘ROCR’ is not available (for R version 3.5.1)
May I know how to solve this? Thank you.
Hi Carmen,
Yes here it says it is only available for R>3.6 https://cran.r-project.org/web/packages/ROCR/index.html.
Try updating the R version and then install the package again same way. It should work.
Hello Team,
I am getting the error when installing the gganimate package, as mentioned below on Linux Ubuntu 20.0..however, I would not install version 3.6.2. What could I do?
> install.packages(“gganimate”, dependencies=TRUE,
+ repos=’https://CRAN.R-project.org/package=gganimate’)
Installing package into ‘/home/sabrina/R/x86_64-pc-linux-gnu-library/3.6’
(as ‘lib’ is unspecified)
Warning in install.packages :
unable to access index for repository https://CRAN.R-project.org/package=gganimate/src/contrib:
cannot open URL ‘https://CRAN.R-project.org/package=gganimate/src/contrib/PACKAGES’
Warning in install.packages :
package ‘gganimate’ is not available (for R version 3.6.3)
I tried this:
install_version (“gganimate”, version = NULL, repos = “http://cran.us.r-project.org”)
But I also couldn’t
Now I got it!
I deleted this package: stringi and installed again
> install.packages(“~/R/x86_64-pc-linux-gnu-library/3.6/gganimate.tar.gz”, repos = NULL, type = “source”)
Installing package into ‘/home/sabrina/R/x86_64-pc-linux-gnu-library/3.6’
(as ‘lib’ is unspecified)
* installing *source* package ‘gganimate’ …
** package ‘gganimate’ successfully unpacked and MD5 sums checked
** using staged installation
** R
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
*** copying figures
** building package indices
** installing vignettes
** testing if installed package can be loaded from temporary location
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (gganimate)
> library(gganimate)
Its cool!
Glad it is fixed now! thanks for sharing
can you just try running like this `install.packages(“gganimate”)`
Whenever I try to attach my dataname (bk) to R, this is the response I get:
The following objects are masked from bk (pos = 7):
How can I resolve this?
Can you give more context? maybe share steps to reproduce so i can try to help.
tried to install pROC or ROCR in R version 4.0.2 but faiiled many times.
install.packages(‘pROC’,repos=’http://cran.us.r-project.org‘)
Installing package into ‘C:/Users/Admin/Documents/R/win-library/4.0’
(as ‘lib’ is unspecified)
Warning: unable to access index for repository http://cran.us.r-project.org/src/contrib:
scheme not supported in URL ‘http://cran.us.r-project.org/src/contrib/PACKAGES’
Error in gzfile(file, mode) : cannot open the connection
In addition: Warning messages:
1: package ‘pROC’ is not available (for R version 4.0.0)
2: In gzfile(file, mode) :
cannot open compressed file ‘C:\Users\Admin\AppData\Local\Temp\Rtmpm4BBAR/libloc_193_ca5a3a5a.rds’, probable reason ‘No such file or directory’
>
Did u try some other mirrors here https://cran.r-project.org/mirrors.html?
I tried to install the package bbmle and i had the following error message
Installing package into ‘D:/Documents/R/win-library/3.4’
(as ‘lib’ is unspecified)
I am trying to install readx1 package using
install.packages(“readx1″,repos=”http://cran.us.r-project.org”)
but it shows error message
Warning message:
“package ‘readx1’ is not available (for R version 3.6.1)”
how can i install it?
i just tried using the Cloud as mirror and it worked for me
`install.packages(“readxl”, repos=”https://cloud.r-project.org/”)`
I am trying to install clues package using
install.packages(“clues″,repos=”http://cran.us.r-project.org”)
but it shows error message
Warning message:
Installing package into ‘C:/Users/Huihui/Documents/R/win-library/4.0’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘clues’ is not available for this version of R
A version of this package for your version of R might be available elsewhere,
see the ideas at
https://cran.r-project.org/doc/manuals/r-patched/R-admin.html#Installing-packages
Seems like the package is removed from CRAN database https://cran.r-project.org/web/packages/clues/index.html. You need to find it somewhere else and manually install.
Thank you for your reply. I see. I will try.
Unable to install below packages .
a. stringr
b. stringi
c. reshape2
d. caret
showing error ina ll packages
> install.packages(“stringr”)
Installing package into ‘/usr/lib64/R/library’
(as ‘lib’ is unspecified)
Warning: unable to access index for repository https://ftp.iitm.ac.in/cran/src/contrib:
cannot open URL ‘https://ftp.iitm.ac.in/cran/src/contrib/PACKAGES’
Warning message:
package ‘stringr’ is not available (for R version 3.5.2)
Ok. This is might be an old post ! but believe it or not, it works with me !
r version is 4.2
Thanks for posting !